The fundamental question directing our research is how the mammalian genome is interpreted in a context dependent manner leading to different cellular states.
Nearly all of the cells of a multicellular organism contain the same genetic information but interpret that information differently, resulting in different cell types.
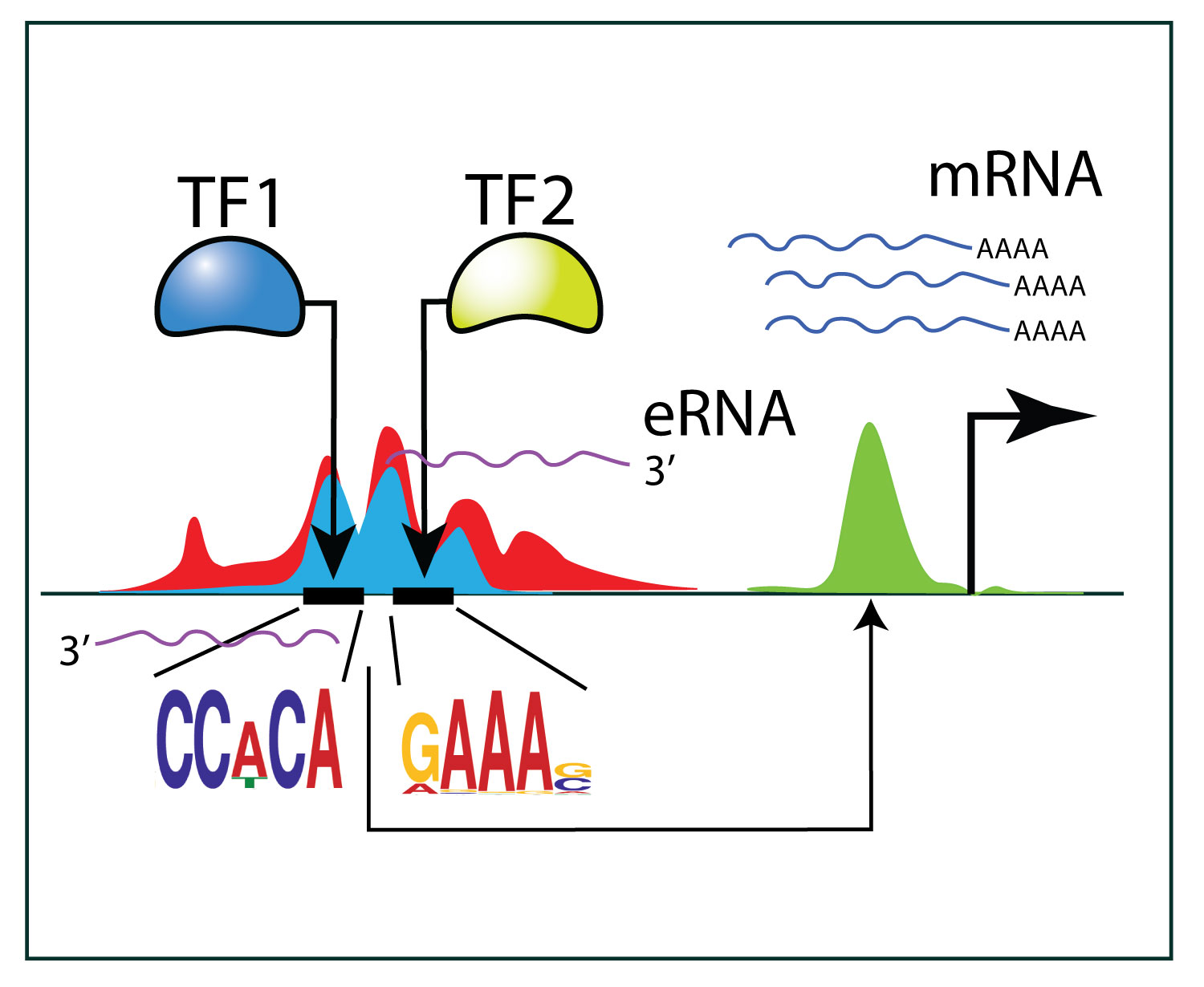
We are interested in sequence specific transcription factors and how they interact with the genetic sequence to co-ordinate different outputs in cell lineage choices and differentiation. We also study cis-regulatory regions in the mouse and human genome, how they interact with transcription factors as well as studying long range interactions of chromatin fibres in the genome.
Our model systems include multiple myeloma and Waldenström’s macroglobulinemia, where we both make use of cell culture models, as well as samples taken directly from patient bone marrow. We hope to use our expertise in transcriptional regulation to reveal the intricacies in disease progression and alteration in cellular states to increasingly aggressive tumour behavior.
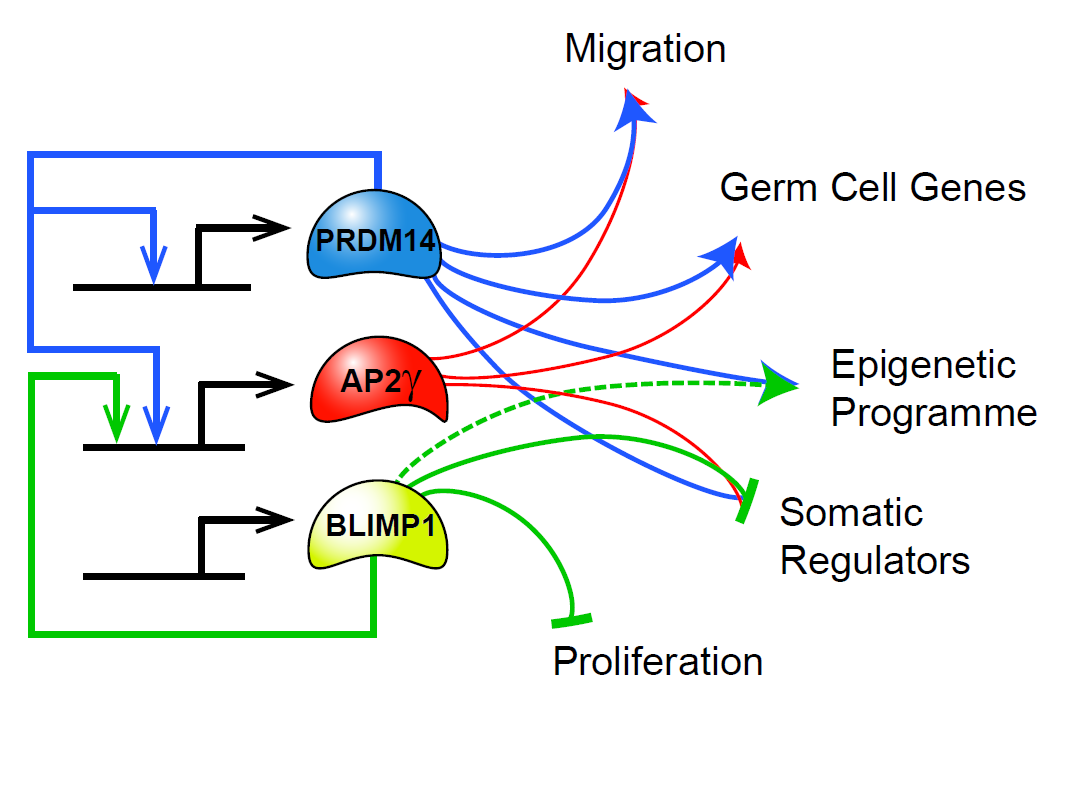
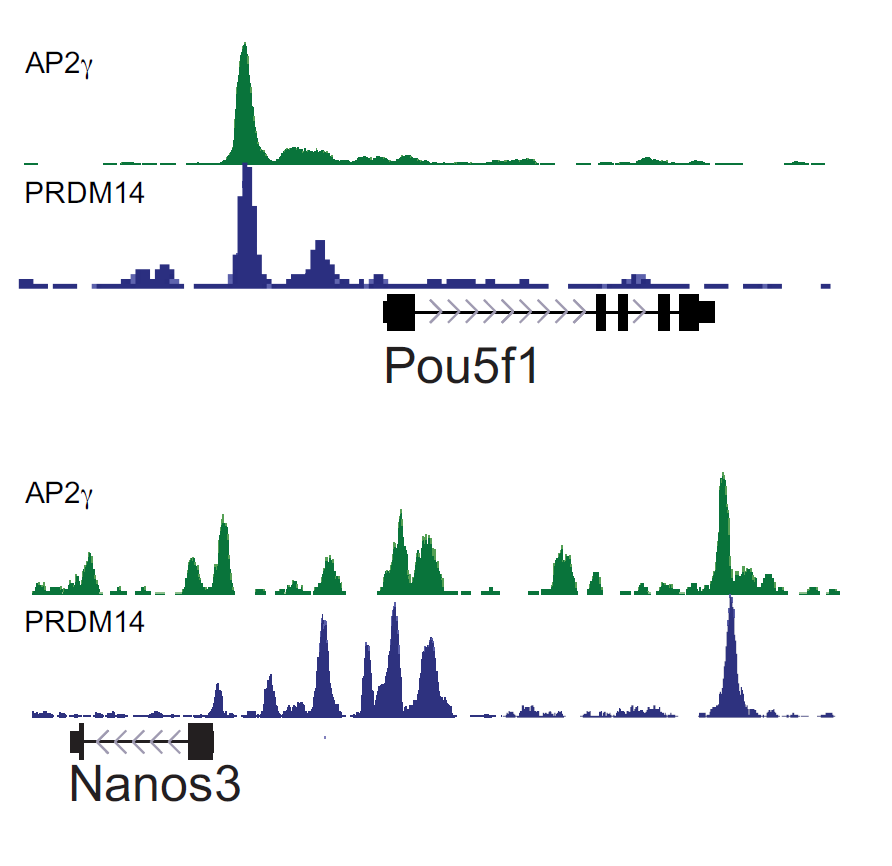
We also study transcriptional regulation of mouse primordial germ cell specification and development using both tissue culture models as well as mouse embryos. We are interested in the transcriptional regulators that drive germ cell fate as well as revealing novel regulators of epigenetic reprogramming occurring in the germ line to prepare the cells for the next generation of an organism.